DNA of dirt: Solving the antibiotic crisis

Using modern technology, students join national research efforts to discover new antibiotics in dirt
By Rhonda Morin
Imagine having a really bad bacterial infection. If doctors knew in a matter of hours—instead of days—what the strain was, they could prescribe an antibiotic to target the nasty bug and begin eliminating it before patients leave the clinic.
Today, there is an answer to those long waits and it all begins with dirt. Clark students are playing a part in the magnificent discovery of cures that lie beneath our feet.
In one cubic foot of soil, more than 30,000 species of bacteria thrive. Buried deep in the guts of those bacteria are some terrific stuff, like antibiotics that help humans fight infections.
For four years, Clark College students have participated in a national study with other colleges and universities—including Yale, University of Connecticut and Washington State University in Pullman—testing soil bacteria for antibiotic activity, as well as identifying bacteria and the chemical structure of antibiotics.
Now, thanks to new technology and an ambitious Clark professor, students are getting the chance to take their research a step further into modern science.
DNA in the field
Hand-held device technology that was first introduced in 2014 enables researchers to test soil samples in the field instead of relying solely on computers housed in laboratories. Clark College students had their first experience with the device during winter quarter 2017 when Clark Biology Professor Roberto Anitori introduced a new science course to the biology department’s curriculum. The device Clark students are using tests soil samples for genetic codes, called deoxyribonucleic acid, more commonly known as DNA.
Anitori has spent his career studying extremophiles—organisms that live in extreme environments. He’s worked in Antarctica and also researched extreme organisms that live and thrive in volcanoes, deep-sea vents, underground water tables and radioactive hot springs.
He’s taught microbiology at Clark since 2008, receiving tenure last year. And he’s an award-winning scientist, having received the Antarctica Service Medal in 2011 from the National Science Foundation for contributing to the continent’s research.
Last September, he launched a new course with the promise of introducing new cutting-edge technology that identifies the genetic makeup of dirt. He caught the interest of potential students by telling them they would be looking for the Ebola and Zika viruses.
The device he’s using in class is revolutionizing how scientists study the formation of antibiotics. It is providing an easy-to-use and cost-effective tool that more researchers can access in order to analyze samples outside of laboratories.
Students collect soil samples on and around Clark’s 101-acre campus in Vancouver’s Central Park during a course they take in the spring. Then they need to analyze the samples. That’s where the new biology course—called Small World Antibiotics Research 2B—comes in.
“We take promising bacteria to find out their entire DNA blueprint—what’s called DNA sequencing,” said Anitori.
It’s similar to spelling out the letters in a piece of writing. Once letters appear, words become clear. Once words are known, then the message is clear.
“DNA sequencing spells out the letters of the genetic blueprint of the microbe, which tells us the information that’s in the DNA that’s telling the microbe to divide now or use these food sources or make this into an antibiotic,” he explained.
The difference now is that scientists and Clark students are using technology that is giving them real-time results. All they need is this hand-held device, a laptop, software and an internet connection.
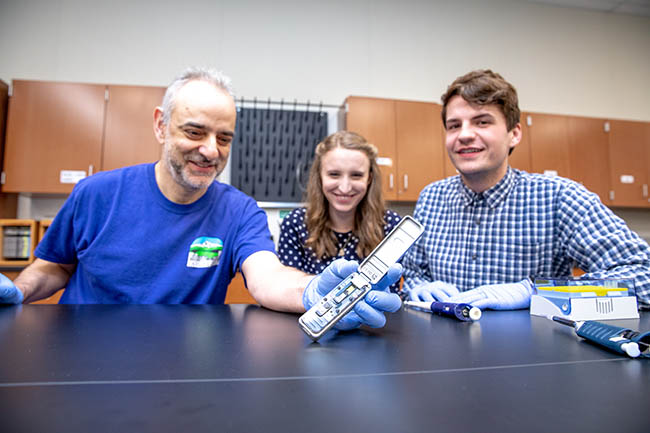
Professor Roberto Anitori demonstrates the MinION to Clark students Georgia Tytler and Havan Sabourin. Photo by Jenny Shadley.
MinION
The device that is changing the way—and where—scientists conduct their work is called a MinION pocket DNA sequencer. Despite the similarity in the name, it’s not related to the computer-animated comedy film that was released in 2015. Rather, it’s manufactured by Oxford Nanopore Technologies from Oxford, United Kingdom.
It’s a tiny device, less than half the size of today’s smart phones. It requires using a micro pipette to slowly drip droplets of liquid into a pinhole-size opening. Then, like a flip-phone, the cover flips into place and the unit connects to a laptop using a universal serial bus (USB) port. Once the start button is pressed, a whizzing sound occurs and the analyzing begins.
“We will get millions and millions and millions of letters from this device. The trick is you then have to put it together,” said Anitori. “The device gives you torn-up pieces of a blueprint. We then use the computer software to put the pieces together.”
This is where Clark students begin their search.
“It’s called mining the genome. We also look at online databases to look for the genes we’re interested in such as the recipes that tell the cells to make an antibiotic,” he said.
Game-changer
There is an antibiotic-resistant crisis going on. Dr. Margaret Chan, former World Health Organization director-general, cautions that resistance to antimicrobials is not a warning for the future. It is a global threat today.
“With few replacement products (antibiotics) in the R&D pipeline, the world is heading toward a post-antibiotic era in which common infections will once again kill… This may even bring the end of modern medicine as we know it,” said Chan in a speech to United Nation member states in New York in 2016.
Bacteria live in soil and make antibiotics. Having the ability to test the soil quickly or in the field gives scientists a time advantage in solving the global antibiotic crisis.
“The great thing about this technology is that it has democratized DNA sequencing,” said Anitori.
It took 15 years and $2.7 billion for scientists to complete the genetic code for the entire human genome, according to the National Human Genome Research Institute. A genome is the full genetic makeup—DNA—present in a cell or organism.
“Now they can do it in a day or two for about $1,000,” said Anitori.

The MinION pocket DNA sequencer is manufactured by Oxford Nanapore Technologies from Oxford, United Kingdom.
Research at Clark
Anitori secured the device for his course by calling Oxford Nanapore Technologies.
“I’ve always believed in my career that if you want something you just have to ask for it,” he said.
So he contacted Oxford Nanapore Technologies and asked them if they partner with higher education institutions. In fact they do, they told him.
“They offered $9,000 in supplies to run the class two times through fall 2019.”
The 14 students who were enrolled in the first course had several opportunities to navigate the four donated devices. No new antibiotic discoveries were made—yet.
What’s next?
Anitori has enough supplies to run the course a second time.
“I got the class off and running and now we’ll have to find alternative sources of funding for beyond 2019.”
This biology course, as well as the soil-gathering course called Small World Antibiotics Research 1 that runs during spring term, are intended to get individuals excited about science and elicit enthusiasm to discover. Having access to modern technology and the opportunity to do research that is relevant to science today prepares students for their careers and personal goals.
Listen to the podcast | Share #penguinchatspodcast